 |
| Address | Staudingerweg 9
55128 Mainz |
| Room | 03-519 |
| Phone | 49 6131-39-2xxxx |
| Fax | 49 6131-39-25441 |
| E-mail | tevrugtm @ uni-mainz.de |
| Homepage | https://michaeltevrugt.de/ |
|
I am assistant professor („Juniorprofessor“) of theoretical physics. Previously, I worked as a postdoctoral researcher at the Department of Applied Mathematics and Theoretical Physics at the University of Cambridge in the group of Prof. Michael E. Cates. I have done a PhD in theoretical physics at the University of Münster in the group of Prof. Raphael Wittkowski, and in parallel a PhD in philosophy of science supervised by Prof. Ulrich Krohs and Dr. Paul M. Näger.
My research interests include (but are not limited to):
- Nonequilibrium statistical mechanics
- Soft and active matter
- Biological physics
- Philosophy of physics
Selected publications:
- Michael te Vrugt, Jens Bickmann and Raphael Wittkowski, Effects of social distancing and isolation on epidemic spreading modeled via dynamical density functional theory, Nature Communications 11, 5576 (2020), https://doi.org/10.1038/s41467-020-19024-0
Selected as Editor’s Highlight
Selected for coverage in Physik Journal
Press reports: WWU Münster, Ruhr-Nachrichten, MyScience, Newsbeezer, Pediatric Radiology, News Bioon, Wemp.App, Osel, Tekk.tv, Heilpraxis.net, Healthcare Hygiene Magazine, Informationsdienst Wissenschaft, Research in Germany, DE24-News, Today Headline, MedicalXpress, Dorstener Zeitung, The Diet World,Avalanches, Wissen.Newzs,Abitur-und-Studium.de, Laborpraxis, Stadt40, Sciende Daily, Münsterland-Zeitung, Deutsches Ärzteblatt
- Michael te Vrugt, Hartmut Löwen and Raphael Wittkowski, Classical dynamical density functional theory: From fundamentals to applications, Advances in Physics 69, 121-247 (2020), https://doi.org/10.1080/00018732.2020.1854965
Press reports: WWU Münster, HHU Düsseldorf, Informationsdienst Wissenschaft
- Michael te Vrugt, Sabine Hossenfelder and Raphael Wittkowski, Mori-Zwanzig formalism for general relativity: a new approach to the averaging problem, Physical Review Letters 127, 231101 (2021), https://doi.org/10.1103/PhysRevLett.127.231101
Press reports: WWU Münster, FIAS Frankfurt, Innovations Report, Pro-Physik
- Michael te Vrugt, How to distinguish between indistinguishable particles, British Journal for the Philosophy of Science (accepted), https://doi.org/10.1086/718495
Covered in BJPS Short Reads
Podcast coverage on Apple Podcasts, Spotify, Audible
- Michael te Vrugt, Tobias Frohoff-Hülsmann, Eyal Heifetz, Uwe Thiele and Raphael Wittkowski, From a microscopic inertial active matter model to the Schrödinger equation, Nature Communications 14, 1302 (2023), https://doi.org/10.1038/s41467-022-35635-1
Selected as Editor’s Highlight
Press reports: WWU Münster
- Stephan Bröker, Jens Bickmann, Michael te Vrugt, Michael E. Cates and Raphael Wittkowski, Orientation-dependent propulsion of active Brownian spheres: from self-advection to programmable cluster shapes, Physical Review Letters 131, 168203 (2023), https://doi.org/10.1103/PhysRevLett.131.168203
Press reports: WWU Münster
More information can be found on my personal homepage: https://michaeltevrugt.de/
How to pronounce my last name: Read „te frücht“ as if it were a German word. (If you’re not German: It’s difficult to explain.)
Die Frist für die Einschreibung in Übungsgruppen ist verstrichen. Vielen Dank für Ihre Teilnahme.
Falls Sie sich nicht eingeschrieben haben, und dennoch an einer Übung teilnehmen möchten, schreiben Sie mir bitte eine Email. Ich teile Sie dann einer Gruppe zu, in der noch Platz ist.
Our group is elucidating with multi-scale simulations how phase separation can provide for specific regulation in biology and how dysregulation of phase separation can lead to disease.
Our group has joined the collaborative research project TRR146 Multiscale Simulation Methods for Soft-Matter Systems for its third funding period with a joint project with Thomas Speck.
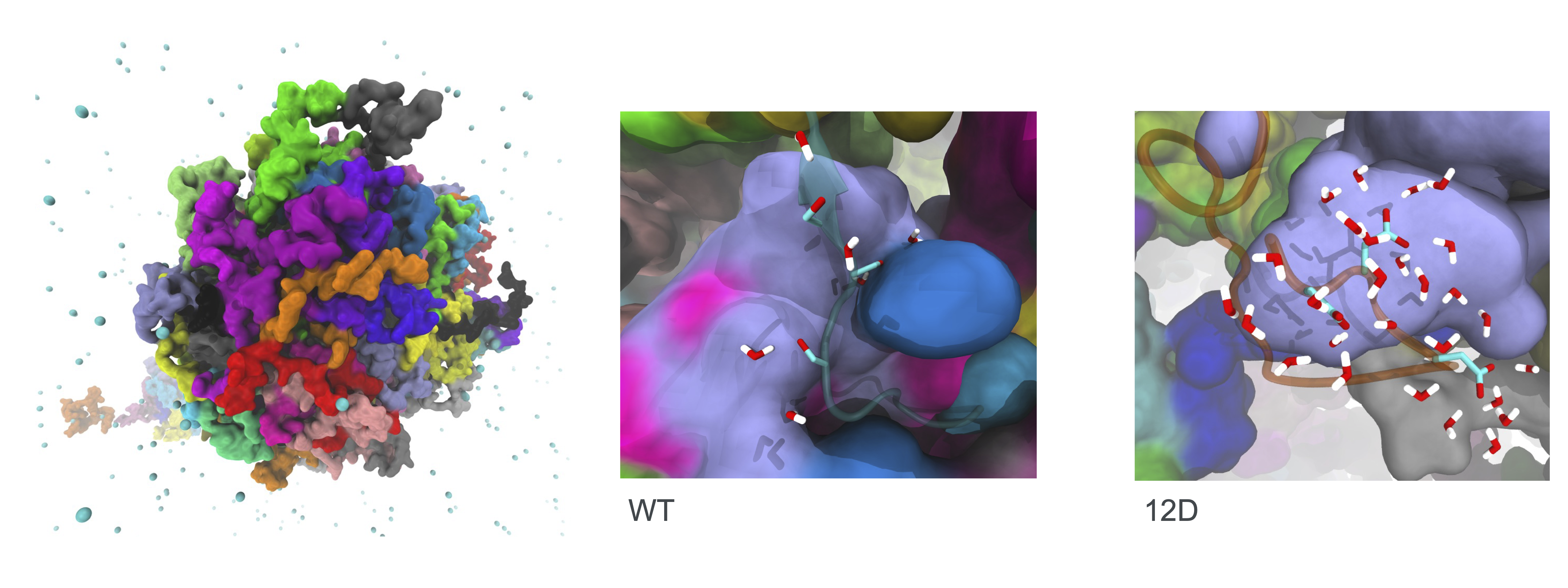
Multi-scale simulations of TDP-43. Left: Phase-separated condensate of the disordered domain of TDP-43 from coarse-grained simulation. Centre: Details of water-binding to sites to serines residues in C-terminus of the disordered domain from atomistic simulations of a TDP-43 condensate. Right: Atomistic simulation of phosphomicking 12D mutant shows more water molecules are bound at sites of phosphomimcking mutations compared to wild-type TDP-43. Figure adapted from Grujis da Silva et al EMBO J, 2022
ReALity Junior Group Leader and IMB Associate Group Leader
Collaborations and Funding
A seminar course on Soft Matter and Biological Physics is offered in the summer term 2019.
Time: Tuesday 14:00-16:00
Place: Newton Raum (01-122)
This course is open to master students (as Master Seminar I or II). Bachelor students with knowledge in statistical physics (Theory 4) are also welcome to attend.
Soft matter is an interdisciplinary field, which combines aspects from physics, chemistry and biology. The range of topics where physicists can contribute is broad and steadily growing. Examples include
- Mechanics and thermodynamics of (bio)polymers
- Biomaterials (membranes, bones, cartilage)
- Physics of microswimmers and active particles
- Microfluidic devices for particle sorting and separation
The purpose of this course is to introduce students into this fascinating field. We will not be able cover all aspects of soft matter, but instead focus on selected topics (see list below). The seminar will provide physical background and present applications in soft matter physics.
Critical reading, analysis and discussion of scientific literature is one of the key aspects of modern science, and therefore we will practice these skills during the first 4 sessions of this seminar course. In particular, we will interactively develop and practice the following work strategies:
- Summarizing articles
- Providing constructive feedback
- Planning and preparing presentations
The scientific part of the seminar is organized as follows:
- Every student is supposed to give an approximately 30 minutes long talk on the selected topic
- Please pick your topic before the beginning of the course by sending an e-mail to Sara Jabbari Farouji and Arash Nikoubashman
- Contact your talk supervisor at least 4 weeks prior to your presentation for material
- Prepare a first version at least 2 weeks before the presentation
- Schedule a practice talk with your supervisor for the week before the presentation
- Upload your presentation to the seafile folder to share it with your colleagues
Literature
Literature will be provided by your talk supervisor.
List of topics
Vorlesung: Freitag 8:00-11:00, HS Kph
Übungen:
Oberassistent: Pascal Wasser (email)
Abgabe rote Kästen Staudingerweg 7 (38: 1+2, 39: 9+4, 40: 5+6, 41: 7+8)
- Gruppe: Mittwoch 12:00-14:00 Uhr (Seminarraum E) Maike Jung
- Gruppe: Mittwoch 12:00-14:00 Uhr (Seminarraum F) Zeki Aydin
- Gruppe: Mittwoch 14:00-16:00 Uhr (Seminarraum A) Annika Stein
- Gruppe: Mittwoch 16:00-18:00 Uhr (Seminarraum F) Andreas Fischer
- Gruppe: Freitag 12:00-14:00 Uhr (Newtonraum) Ardit Krasniqi
- Gruppe: Freitag 12:00-14:00 Uhr (Seminarraum D) Dennis Layh
- Gruppe: Freitag 12:00-14:00 Uhr (Seminarraum F) Niklas Pfeifer
Übungsblätter
Für die Klausurzulassung müssen 50% der Aufgaben bearbeitet, 50% der Punkte erreicht und einmal vorgerechnet werden.
Übungsblätter erscheinen online im JGU reader. Zugang über Uni-Account für alle über Jogustine angemeldeten Teilnehmer der Vorlesung.
Literatur und Material
Skripte:
Online:
Bücher:
- P. van Dongen, Einführungskurs Mathematik und Rechenmethoden
- W. Nolting, Theoretische Physik Bd. 1 (erstes Kapitel)
- S. Großmann, Mathematischer Einführungskurs für die Physik
- K.-H. Goldhorn und H.-P. Heinz, Mathematik für Physiker 1
- C. Lang und N. Pucker, Mathematische Methoden in der Physik
- M.L. Boas, Mathematical Methods in the Physical Sciences
- und viele mehr
A seminar/lecture course on Biological Physics was offered in the summer term 2018.
Time: Thursday 2pm - 4pm
Place: Galilei-room (Staudingerweg 9, First floor)
It is open to master students (as Master Seminar I or II). Bachelor students with knowledge in statistical physics (Theory 4) are also welcome to attend.
Biological physics is an interdisciplinary field where methods and tools from physics are applied to problems in biology. The range of topics where physicists can contribute is broad and steadily growing. Examples are
- Mechanics and thermodynamics of proteins
- Biomaterials (membranes, bones, cartilage)
- Cell mechanics and thermodynamics of cellular processes
- Bioimaging and high resolution microscopy
- Physics of living tissues (including: physics of cancer)
- Biological networks
- Neuroscience
- Evolution
The purpose of the course is to introduce students into this fascinating field. We will not be able cover everything, but instead focus on selected aspects depending on the interests of the participants. The lectures will provide physical background (e.g., phase transitions, polymer physics, stochastic processes), and the seminar talks will present applications in biological physics. The topics of the talks are assigned to the students on April 19th.
Literature
- Biophysics, R. Glaser, Springer (available in the PMC library)
- Physics of Life, IFF spring school 2018 Lecture Notes, available online here.
- Biophysics: Searching for principles, W. Bialek, Princeton Univ. Press, preprint available here.
- Molecular Biology of the Cell B. Alberts, A. Johnson, J. Lewis, D. Morgan, M. Raff, K. Roberts, Garland Science Publications (provides biology background, available in the PMC library)
Further literature and the slides of the presentations will be made available to participants here. Please log in with your ZDV credentials.
List of topics
Date/Contact
|
Topic
|
Literature
(for example) |
| 19.4, 26.4, 3.5. |
Introductory lectures: Some physics background |
|
| 17. 5. |
Methods: Experimental (superresolution microscopy, scattering) |
[2] A1-A3 or A4,A5 |
| G. Settanni |
Methods: Simulations (biomolecular simulations)
|
[2] A9, A10 |
| 24. 5. |
Molecules - DNA: Sequence and alignment |
tba |
| P. Virnau |
Molecules - DNA: Molecular organization and chromatin |
[2] B5 |
| 1. 6. |
Molecules - Proteins: Structure and crystallography |
[2] A4, A5 |
| F. Schmid |
Molecules - Proteins: protein folding |
[3] Chap. III.A |
| 7. 6. |
Cell components: Membranes |
[2] C1 |
| F. Schmid |
Cell components: Cytoskeleton |
[2] C3, C6 |
| 14. 6. |
Cellular organization: Cell states and Turing patterns |
[3] Chap III.C, |
| P. Virnau |
Cellular organization: Tissue growth
|
[2] E6 |
| 21. 6. |
Neurons: Ion channels and neuronal dynamics |
[3] Chapter III.B |
| F. Schmid |
Neurons: Neural networks |
[3] Chapter III.D |
| 28. 6. |
Transport: Hydrodynamics in biology |
[1] Chap 3.7 |
| T. Speck |
Transport: Motility of cells and bacteria |
[3(final)] Chap 4.2 |
| 5. 7. |
Evolution |
[2] F2, F7 |
| F. Schmid |
Systems theory |
[1] Ch. 5, [3] Ch. IV |







